Validation and Detection of Some New Rice Hybrids to Salt Stress Using Various Statistical Analysis Methods
| Received 02 Nov, 2024 |
Accepted 09 Jan, 2025 |
Published 10 Jan, 2025 |
Background and Objective: Rice (Oryza sativa L.) is crucial for Egypt’s agricultural sustainability and food security, but its cultivation is hindered by salinity stress, water scarcity and rising temperatures. These challenges are pronounced in salt-affected areas, which account for 50-60% of rice-growing regions. The present investigation was conducted to realize the feasibility of correlation, path coefficient and principal component analysis methods using three-line systems in rice (CMS, maintainer and restorer lines) for newly hybrid rice combinations under salinity conditions. Materials and Methods: The experiment was carried out during the 2022 and 2023 growing seasons at (RRTC) Farm, Sakha, Kafr El-Sheikh as well as Elsie station, Egypt. Twelve F1 hybrids obtained from crossing three CMS lines as females with four restorer lines were evaluated along with the parents for genotypic variation. Analysis of variance was measured among rice genotypes for all traits. Results: The results revealed that grain yield per plant was highly significant positively correlated with each of panicle length, panicle weight and potassium content under normal conditions, while was highly significant positively correlated with panicle weight, spikelet’s sterility percentage, panicles per plant and thousand-grain weight under the salinity condition. The path coefficient analysis showed that the trait with the most direct positive effect on grain yield from endogenous variables appeared from the group of number of panicles (0.777 and 0.780), followed by panicle length (7.436 and 0.502) under normal and salinity conditions, respectively. The best genotypes based on salinity tolerance including the first cluster have Giza179 and GZ9399 and the crosses GAB-A/PR-6 and IR69625A/GZ9399 with best selected traits under salinity conditions such as panicle weight and grain yield. Conclusion: The promising hybrid combinations; G46A/PR6, G46A/PR5, GAB-A/PR6 and IR69625A/Giza179 could use as a good crosses.
| Copyright © 2025 Zayed et al. This is an open-access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
INTRODUCTION
Rice (Oryza sativa L.) has a very important role and strategic way in Egypt’s agricultural sustainability and raising its yield is considered a vital option to secure food security and sustainability. The salinity problem involving saline soil and water is the main grand challenge rice cultivation and production in which salt-affected soils represent 50-60% out of total rice cultivated area. Rice is a strategic crop and a main mail in several countries, including Egypt, whereas it is cultivated in salt-affected coastal areas as a reclamation crop despite its sensitivity. Rice is especially well-relevant for the regions affected by waterlogging, as the standing water helps alleviate and leach out salts from the soil whereas the root zone drains1. Interestingly, the cultivation and production of rice in Egypt suffer several constraints like salt stress, high temperature and low fresh water supply, with about 30-35% of irrigated soil of the delta classified as salt-affected soils2.
Researchers interested in rice at Egypt’s Rice Research and Training Center (RRTC) released several varieties namely; Sakha104, Giza178 and Giza179 as well as EHR1 and GZ9399 to cope with those threats in terms of salinity and low water supply3. The most of salt-affected contain various soluble salts involving cations and anions; Ca+2, Mg+2, Na+ and SO42–, Cl–, HCO3–, can hinder crop production, particularly when the salinity level of more than 4 which is sufficient to decline yields particularly rice since its salinity threshold limit is 3. The pH of saline soils typically ranges from 7-8.5 but can vary in saturated conditions, potentially inducing sharp crop loss4. Salt stress negatively affects rice by slowing growth, diminishing metabolism and restricting water and nutrient uptake. Furthermore, high sterile spikelets, particularly inferior ones, under salt stress resulted in a significant decline in rice grain yield.
As previously cited, hybrid rice had high heterosis compared to inbred ones which enabled it to surpass the inbred rice varieties regarding the growth and grain yield. Furthermore, the high heterosis of hybrid rice might be induced by high abiotic stress tolerance such as salinity, heat and drought stresses. Zayed et al.5 evaluated some rice hybrids compared to inbred rice under salt stress and they found that the tested hybrids had more salt tolerance than inbred owing to their high physiological and morphological heterosis. Ramadan et al.6 used PCA to classify some rice genotypes into different clusters with considering their salinity tolerance and salt gene. They found that PCA analysis classified the tested rice genotypes into four groups based on their tolerance and subspecies as well saltol gene.
Under aerobic conditions, using correlation, path coefficient analysis as well as PCA in terms of Eigen value analysis was very effective in detecting the genetic diversity among tested rice genotypes. In addition, the various used statistics analyses showed trustful classification of tested rice genotype to different clusters based on its drought withstanding indicating the validation of Giza179, Giza178, EHR1 and Gz9399 to aerobic conditions3. Understanding the genetic behavior of salinity tolerance is a major concern in producing new genotypes suitable for salt-affected areas in Egypt. In the breeding program, the knowledge of genetic variability for evolving high grain is important under salt stress. Genetic advancement is more helpful in predicting the gain under selection than heritability estimates alone7. The present investigation aimed to study the path coefficient, principal components and cluster analysis of three diverse CMS lines, four restorer lines and their hybrid combinations for yield and some other traits in hybrid rice under normal and saline soil. Therefore, the wider goal of the current attempt is nominating the most alt-tolerant rice hybrid which copes with salinity and climate change stresses.
MATERIALS AND METHODS
Study area and duration: The present investigation was conducted at the Experimental Farm of Rice Research and Training Center (RRTC), Sakha, Kafer El-Sheikh and El-Sirw Station, Egypt, during the two rice successive seasons 2022 and 2023.
Materials parentage: The genetic materials used in this investigation involved three CMS lines of rice obtained from different sterile sources of Wild Abortive (WA) and Gambiaca (three-line system). These lines were; IR69625A, G46A and GAB-A which were used as female parents. In addition, four Egyptian tester lines viz., Giza179, PR5, PR6 and GZ9399 were used as testers shown in Table 1.
Methods: The grains of 12 F1 hybrids were obtained in the growing season of 2022, according to line×tester mating design (3×4) involving the three CMS lines (lines) and four restorer lines (testers). The obtained grains of the 12 F1 hybrids in addition to their parents were evaluated in the 2023 growing season under both normal and saline soil conditions. The experiment was sown in the nursery during the first week of May. Seedlings of the parental lines and their F1 hybrids, of 30 days of age, were individually transplanted in a randomized complete blocks design with three replicates. Each replicate consisted of 3 rows. The rows were five meters long with 20 cm between rows comprising 25 hills each for a single plant. All agricultural practices were made according to rice recommendations. The data were recorded on an individual plant basis for parents and F1 generation.
| Table 1: | Cytoplasmic male sterile lines and tester lines used for this study | |||
| Genotypes | Cytoplasmic source pedigree | Grain type | Origin |
| PTGMS lines (female) CMS lines | |||
| IR69625A | Wild abortive (WA) CMS line | Medium grain | IRRI |
| G46A | Gambiaca | Short grain | China |
| GAB-A | Gambiaca | Medium grain | Egypt |
| Tester lines (male) restorer line | |||
| Giza179 | GZ1368-5-S-5/GZ6296-12-1-2-1-1 | Short grain | Egypt |
| PR5 | Giza 178/GZ6296-12-1-2-1-1 | Short grain | Egypt |
| PR6 | Giza 178/GZ6296-12-1-2-1-1 | Short grain | Egypt |
| GZ9399 | Giza 178/IR65844-29-1-3-1-2 | Short grain | Egypt |
| Table 2: | Some chemical and physical analysis of experimental sites during the 2022 season | |||
| meq/L | ||||||||||
| Soil | EC | pH | Ca+2 | Mg+2 | Na+ | K+ | CO32– | HCO3– | Cl– | SO42– |
| Normal | 2.07 | 7.59 | 2.98 | 1.12 | 14.96 | 0.47 | 2.17 | 1.34 | 13.35 | 5.36 |
| Salinity | 8.13 | 8.46 | 12.9 | 6.1 | 65.23 | 0.35 | 6.82 | 7.36 | 61.29 | 12.33 |
Soil analysis: The soil samples were taken from the experimental farm at El-Sirw Agriculture Research Station (salinity) and Sakha Agriculture Research Station (normal) and before conducting the experiments, soil samples from two sites were taken from depths 0-30 cm, all samples were then air dried and prepared for chemical analysis. The chemical analysis was carried out using the soil extract 1:5 to estimate the soluble anions, cations and total dissolved salts (TDS). Soil saturation extract was measured according to Wolf8. The electrical conductivity (EC) was measured in the extract of the soil-saturated past9,10. Some chemical traits of the soil of the experimental sites at El-Sirw and Sakha stations in the 2022 season were given in Table 2.
Observations were recorded on the following traits: The data were recorded on five randomly selected plants from each replication for various quantitative traits studied viz., days to heading, plant height, panicles per plant, panicle length, spikelet’s fertility (%), 1000-grain weight, panicle weight and yield per plant. Sodium was determined by flame photometry technique. Sodium content (%)11 and potassium content (%) were recorded via method of Dewey and Lu12. Potassium was also determined by using the flame photometry method.
Statistical analysis: Phenotypic correlation coefficients were assessed by the standard procedure according to Miller et al.11. The path analysis was performed as described by Dewey and Lu12 method. The statistic pseudo-F statistic and the pseudo-T213 were tested to address the numbers of clusters using SAS version 9.2 software (SAS Institute, 1996, Cary, NC). Furthermore, the patterns of morphological variation, (PCA) were performed. The clustering of various tested rice genotypes into similar groups was carried out by Ward’s hierarchical algorithm based on squared Euclidean distances.
RESULTS AND DISCUSSION
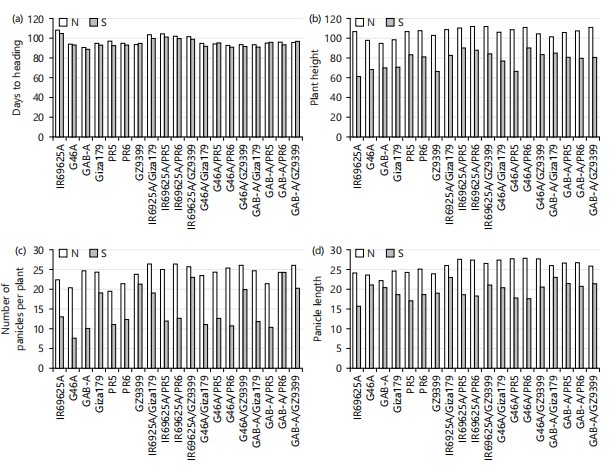
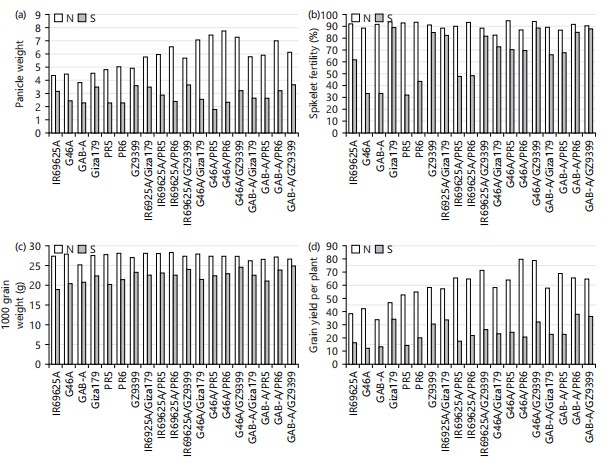
Mean performance: Mean performance of seven parental lines (three CMS and four restorers) and their 12 hybrid combinations of line×tester for the nine studied agronomic traits i.e., days to heading (day), plant height (cm), number of panicles plant, panicle length (cm) Fig. 1(a-d) and panicle weight (g), spikelet fertility (%), 1000-grain weight (g) and grain yield per plant (g) are presented in Fig. 2(a-d), respectively.
|
The results indicated that the mean performance for the studied traits varied from one combination to another. Previous results found that field performance for all agronomic traits showed significant differences among the 64 crosses and their pollen parents at p<0.05 and 0.01 probability levels. Early maturity is a desirable trait in rice. For days to heading, among parents, GAB-A was the early maturing line under both conditions normal and saline with values of 90.33 and 88.67 days. The highest mean values belonged to IR69625A under both conditions with values of 107.33 and 105.0 days, respectively. For the hybrid combinations, G46A/PR6 gave the lowest mean values (92.0 and 91.0 days) under both conditions, respectively. While, the highest mean values for this trait were obtained from IR69625A/PR5 under normal and salinity conditions with values of 103.67 and 101.0 days, respectively in Fig. 1a. For plant height the results showed that the genotype, GAB-A gave the lowest mean value 94 cm under normal condition while IR69625A were the shortest plants under saline soil condition with 61 cm followed by G46A/PR5 with 66.33 cm. On the other hand, two crosses IR69625A/PR6 and IR69625A/GZ9399 gave the highest value of plant height 111 cm under the normal condition shown in Fig. 1b. Meanwhile, two crosses G46A/PR6 and IR69625A/PR6 gave the highest value of plant height 89.67 cm under salinity condition. It is worth note noting that most of the F1 mean values were directed toward the tall parents. These, in turn, suggested that tallness was dominant under saline soil conditions14. These results agree with Zayed et al.15 and disagree with Faiz et al.16, they mentioned that both the CMS lines reduced the plant height of their respective F1 hybrids. For panicles per plant, data showed that the most desirable mean values towards this trait were obtained from the parents GAB-A (24.67 ) under normal conditions, while the best parent under salinity conditions was GZ9399 (21.33) followed by Giza179 (19.00).
|
Furthermore, the most desirable mean values of panicles per plant were obtained from the crosses, IR69625A/Giza179 and IR69625A/PR6 under normal conditions; GAB-A/PR6 and IR69625A/GZ9399 under salinity conditions. Meanwhile, the lowest values of panicles per plant were detected by the parents, PR5 and G46A under normal conditions and GAB-A and G46A under salinity conditions in Fig. 1c. For crosses GAB-A/PR5 under normal conditions and GAB-A/PR5 gave the lowest mean value under salinity stresses. Concerning panicle length, the results indicated that the most desirable mean values were obtained from the parents, PR6 (25.03 cm) and the crosses G46A/PR6 (27.83 cm) followed by G46A/GZ9399 (27.57 cm) under normal, GAB-A/Giza179 (23.00 cm) under salinity condition. On the other hand, the lowest values obtained from the parent, GAB-A under normal and IR69625A under saline soil conditions are shown in Fig. 1d.
Regarding panicle weight, the highest mean values were observed for parents PR6 under normal conditions (5.03) and GZ9399 under salinity conditions (3.60). The lowest mean values were obtained by GAB-A under normal condition (3.80) and GAB-A along with PR5 and PR6 under salinity conditions (2.30) in Fig. 2a. On the other side, among hybrids the highest mean values were observed for G46A/PR5 and G46A/PR6 under normal condition (7.43 and 7.73, respectively), in addition, IR69625A/GZ9399 and GAB-A/GZ9399 under salinity (3.63). Meanwhile, the lowest mean values were recorded for the hybrid IR69625A/Giza179 under normal and G46A/PR5 under salinity conditions (5.80 and 1.80, respectively). For spikelet’s fertility percentage, among parents the highest mean value and most desirable under normal and salinity conditions were scored by Giza179 (93.67 and 89.19%, respectively); while the lowest mean value was obtained by G46A (88.63%) under normal conditions, PR5 (31.69%) under salinity stress in Fig. 2b. On the other side, among hybrids, under normal condition, G46A/PR5 scored the highest mean values (94.63%); furthermore, G46A/GZ9399 and GAB-A/GZ9399 exhibited the highest mean values under salinity conditions (88.15 and 87.88%, respectively). For 1000-grain weight, as in Fig. 2 among parents, under normal conditions, the highest mean values were scored by PR6 (28.00 g) and GZ9399 (23.33 g) under salinity conditions, while, the lowest mean value was obtained by GAB-A under normal conditions and IR69625A under salinity condition. Among hybrids, the highest mean values were scored by the hybrids IR69625A/PR6, IR69625A/Giza179 and IR69625A/PR5 under normal, while GAB-A/GZ9399 was the best cross under salinity conditions. Meanwhile, the lowest mean values were recorded by GAB-A/Giza179 under normal conditions and GAB-A/PR5 under salinity stress in Fig. 2c. For grain yield per plant, data indicated that among parents the highest mean values were obtained by GZ9399 (58.27) under normal while Giza179 and GZ9399 were given the highest grains weight for the single plant under salinity conditions (34.1 and 30.77 g) in Fig. 2d. Meanwhile, the lowest mean values were exhibited for GAB-A (33.90 g) under normal condition furthermore, two parents G46A and GAB-A gave the lowest values under salinity condition (12.2 and 13.17 g, respectively). Among hybrids, the highest mean values under normal conditions were scored by the hybrids G46A/PR6 (79.61 g) and the cross GAB-A/PR6 under salinity conditions (38.00 g). Meanwhile, the lowest mean values exhibited under normal conditions were scored by the hybrids IR69625A/Giza179 (57.17 g) and the cross IR69625A/PR5 under salinity conditions (17.47 g). The same trend was found by El Mowafi et al.17 they reported that the mean performance of grain yield of best promising hybrids under saline conditions ranged from 3.22 ton/fed, for IR69625A/PR1 to 3.34 ton/fed, for IR69625A/GZ5121R.
| Table 1: | Phenotypic correlation coefficients among the studied traits were estimated under normal and salinity conditions | |||
| Traits | DTH | PLH | NOP | PL | PW | STR | TGW | |
| PLH | N | 0.51* | - | - | - | - | - | - |
| S | -0.05 | - | - | - | - | - | - | |
| NOP | N | 0.15 | 0.31 | - | - | - | - | - |
| S | 0.17 | 0.03 | - | - | - | - | - | |
| PL | N | 0.11 | 0.69** | 0.46* | - | - | - | - |
| S | -0.23 | 0.26 | 0.24 | - | - | - | - | |
| PW | N | -0.1 | 0.60** | 0.46* | 0.94** | - | - | - |
| S | 0.35 | -0.01 | 0.82** | 0.33 | - | - | - | |
| STR | N | 0.06 | -0.07 | 0.02 | -0.22 | -0.2 | - | - |
| S | 0.11 | -0.01 | 0.78** | 0.29 | 0.70** | - | - | |
| TGW | N | 0.49* | 0.49* | -0.11 | 0.39 | 0.26 | 0.03 | - |
| S | -0.05 | 0.47* | 0.71** | 0.39 | 0.50* | 0.67** | - | |
| GYP | N | -0.05 | 0.67** | 0.45 | 0.87** | 0.85** | -0.17 | 0.21 |
| S | 0.03 | 0.09 | 0.87** | 0.37 | 0.69** | 0.90** | 0.74** | |
| *Correlation is significant at the 0.05 level, **Correlation is significant at the 0.01 level. Where, DTH: Days to heading, PLH: Plant height in centimeters, NOP: Number of panicles per plant, GYP: Grain yield per plant, TGW: 1000-grains weight, STR: Spikelets fertility (%), PL: Panicle length, PW: Panicle weight (g), Na+: Sodium content (%), K+: Potassium content (%), N: Normal and S: Salinity | ||||||||
Phenotypic correlation coefficient: For plant height, a significant and highly significant positive correlation was obtained for days to heading, panicle length, panicle weight, grain yield and potassium content under normal conditions. Meanwhile, the plant height trait as indicated in Table 3 had a significant positive correlation with thousand grain weight under normal and salinity conditions. Days to heading expressed a significant positive correlation with plant height and thousand-grain weight under normal conditions. For several panicles per plant, it was observed that the number of panicles per plant had a significant and highly significant positive correlation with thousand-grain weight, panicle length, panicle weight, spikelet’s sterility percentage and grain yield per plant under normal conditions. Also, the current number of panicles per plant had a highly significant positive correlation with panicle weight under salinity conditions. Regarding panicle length, a significant and highly significant positive correlation was obtained for panicle length with plant height, number of panicles per plant, plant weight, grain yield per plant and
potassium content under normal conditions. For panicle weight, panicle weight possessed significant and highly significant positive correlation with plant height, panicle length, number of panicles per plant and grain yield per plant undernormal condition. Furthermore, there was a significant and highly significant positive correlation with spikelet’s sterility percentage, thousand-grain weight and grain yield per plant under salinity conditions. The trait of spikelets fertility percentage had a significant and highly significant positive correlation with panicle weight, number of panicles per plant, thousand-grain weight and grain yield per plant under salinity conditions, indicating the high differences among the parent in the crosses and the heterosis with high yielding controlling by spikelet’s sterility percentage. As indicated here sterility in terms of panicle fertility plays a direct role in yield formation under salinity conditions. Thereby, the rice plant breeder working on yielding rice under salt stress should pay attention to improving panicle characteristics, particularly, panicle fertility and reducing panicle sterility as much possible. Moreover, spikelet sterility percentage was not significant under normal conditions with any traits. Regarding thousand grain weight, it had a significant and highly significant positive correlation with days to heading and plant height under normal conditions. At the same time, it was a significant and highly significant positive correlation with plant height, number of panicles per plant, panicle weight, spikelets sterility percentage and grain yield per plant under salinity condition. Again, panicle characteristics including 1000-grain weight in the terms effectively improved yield under salt stress. Regarding potassium content, the potassium leaf content significantly positively correlated with panicle length and grain yield per plant under normal conditions. On the other hand, a significant negative correlation was observed with plant height under salinity conditions.
Data in Table 3 revealed that grain yield per plant was significant and positively correlated with each plant height18,19, panicle length20,21, panicle weight22,23 and potassium content24 under normal conditions. Furthermore, the grain yield per plant was highly significantly positively correlated with each panicle weight22, spikelet’s sterility percentage, panicles per plant20,21 and thousand-grain weight under the salinity condition. The entire information related to the interlink of plant traits such as rice grain yield with yield attributes is an important key to the breeder for doing sense improvement in complex quantitative traits like grain yield for which direct selection is not much acting. Hence, a corresponding analysis was formed to detect the direction of selection and number of traits in which they could be used for improving rice grain yield.
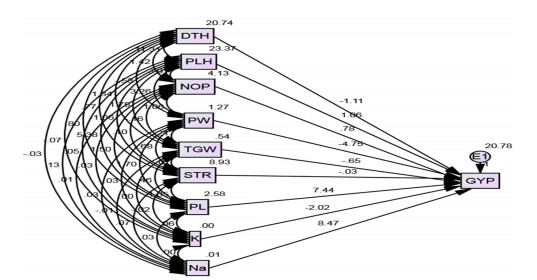
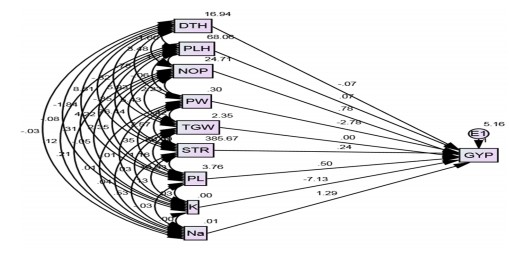
Path coefficient analysis: It is well known that path analysis classifies the correlation to direct and indirect impacts and assesses the relative importance of the factors included. The data of interlinks among various characteristics is too important when the selection for their vital contribution to improvement is the main goal of the plant breeder. If two desirable traits are associated, selection for one will automatically be good enough to make meaningful improvements to the other3. The structure path analysis implemented the use of AMOS software to see the direct and indirect influence of the investigated traits on grain yield under normal and salinity environments. Figure 3 and 4 involved the findings of path analysis diagrams in both studied ecologies. The parameters with the most direct positive impact on grain yield from endogenous variables were clarified from the group of panicle numbers (0.777 and 0.780), followed by panicle length (7.436 and 0.502) under normal and salinity conditions, respectively. The current findings provide the importance of the number of panicles and panicle length for raising rice grain yield, particularly under stresses such as salinity. These results did agree with those reported by Kiani and Agahi25 and Zarbafi et al.26 as well as Zayed et al.27.
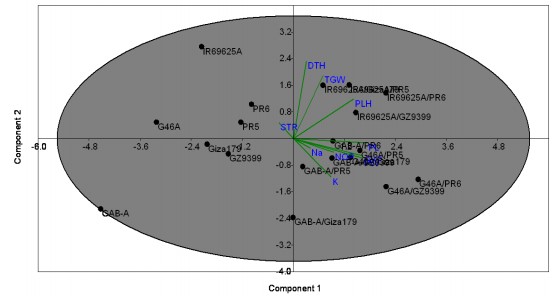
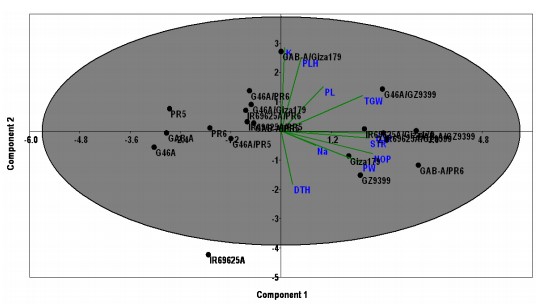
Principal component analysis: The Principal Component Analysis (PCA) was assessed to find out the genetic linkage and relations among the presently tested rice hybrids and genotypes. The PCA analysis divided the studied genotypes into clearly main four clusters based on their origin and salinity withstanding under normal conditions (Fig. 5), the first cluster included the genotypes Giza179 and
|
|
|
|
| Table 4: | Eigenvalue and principal component based on studied traits | |||
| Normal | Salinity | |||||
| PC | Eigenvalue | Variance (%) | Cumulative (%) | Eigenvalue | Variance (%) | Cumulative (%) |
| 1 | 4.049 | 40.49 | 40.49 | 4.275 | 42.75 | 42.75 |
| 2 | 1.789 | 17.89 | 58.38 | 2.004 | 20.04 | 62.79 |
| 3 | 1.326 | 13.26 | 71.64 | 1.149 | 11.49 | 74.28 |
| 4 | - | - | - | 1.017 | 10.17 | 84.44 |
GZ9399 in addition to GAB-A and the cross GAB-A/Giza179. The second cluster involved the four genotypes IR69625A, G46A, PR-5 and PR-6. While the third cluster contains the crosses of the CMS line GAB-A and G46A. Meanwhile, all other crosses from the CMS line IR69625A come from the same cluster approximately. On the other hand, under salinity conditions Fig. 6, the genotypes classified based on salinity tolerance in which the tolerant genotypes listed in the same cluster with good traits could be used in a breeding program. The first cluster has Giza179 and GZ9399 and the crosses GAB-A/PR-6 and IR69625A/GZ9399 with best-selected traits under salinity conditions such as panicle weight and grain yield.
The first three principal components scored the maximum Eigenvalues which were more than one of 40.49% (PC1),17.89% (PC2) and 13.26% (PC3) explaining the cumulative variation of 71.46% of the total variation among the studied genotypes under normal condition. Moreover, under salinity conditions, the first four principal components scored the maximum Eigenvalues which were more than one of 42.75% (PC1), 20.04% (PC2), 11.49% (PC3) and 10.17% (PC4) explaining the cumulative variation of 84.44% of the total variation among the studied genotypes under salinity condition (Table 4). These results indicated the ability of PCA analysis to distinguish the tolerant genotypes from the susceptible ones based on the studied traits used in the current study.
Cluster analysis
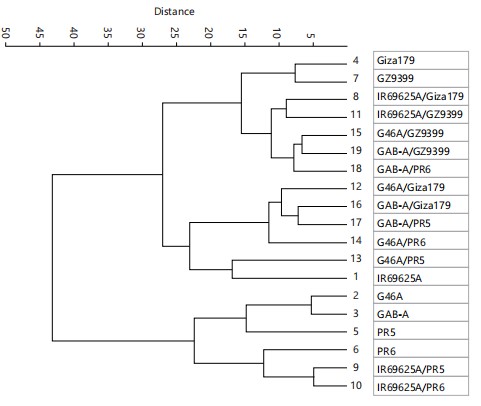
Estimates of cluster analysis under salinity conditions: Hierarchical cluster analysis based on the vegetative stage was presented in Fig. 7. From analyzing the dendrogram at distance 28, it could be observed that there are two main distinct groups. The first group comprises four genotypes with more substantial similarities: IR69625A, G46A, GAB-A and Giza179. Within this group, genotypes G46A and Giza179 show the closest relationship, joining at a low distance (8). Genotype GAB-A joins this pair at a low distance (13), followed by genotype IR69625A at a higher distance (19). This structure suggests that while all four genotypes share some common characteristics, there’s a more substantial similarity between G46A and Giza179 genotypes, with GAB-A being the next closest and IR69625A being somewhat more distinct within the group.

|
The larger group (second group) encompasses the remaining 15 genotypes (PR5, PR6, GZ9399, IR69625A/Giza179, IR69625A/PR5, IR69625A/PR6, IR69625A/GZ9399, G46A/Giza179, G46A/PR5, G46A/PR6, G46A/GZ9399, GAB-A/Giza179, GAB-A/PR5, GAB-A/PR6 and GAB-A/GZ9399). Analyzing the dendrogram, the second group can be divided into two distinct subgroups: Subgroup 1: This subgroup includes twelve genotypes, PR5, PR6, GZ9399, G46A/Giza179, IR69625A/Giza179, IR69625A/PR5, IR69625A/PR6, IR69625A/GZ9399, G46A/PR5, GAB-A/Giza179, GAB-A/PR5 and GAB-A/PR6. Subgroup 2: This subgroup consists of two genotypes, G46A/PR6 and G46A/GZ9399. Analyzing subgroup 1 divided into two groups at a distance of about 14 to sub-subgroup 1 contains five genotypes, PR5, PR6, GZ9399, GAB-A/Giza179, G46A/Giza179 and sub-subgroup 2 contains eight genotypes IR69625A/Giza179, IR69625A/PR5, IR69625A/PR6, IR69625A/GZ9399, G46A/PR5, GAB-A/PR6, GAB-A/GZ9399 and GAB-A/PR5. Sub-subgroup 1 showed that genotypes (PR5 and PR6) and (GZ9399 and GAB-A/Giza179) were the closest pair, joining at the lowest distance (around three units on the vertical axis). Genotype G46A/Giza179 joins the cluster at a distance of about nine units, which is more closely related to the (GZ9399 and GAB-A/Giza179) than genotypes (PR5 and PR6). The sub-subgroup 1 showed that genotypes (G46A/PR5 and GAB-A/PR6) show the closest relationship in the groups, joining at a low distance (4) next genotype GAB-A/GZ9399 then joins this pair at a low distance of about (6) and genotype GAB-A/PR5 one of the more distinct genotypes, joining the larger cluster (G46A/PR5, GAB-A/PR6, GAB-A/GZ9399) at a high distance about (8). Genotypes (IR69625A/PR5 and IR69625A/PR6) show the closest relationship, joining at a low distance about (5); next, genotype IR69625A/GZ9399 joins this pair at a low distance (7). Genotypes IR69625A/Giza179 follow sub-subgroup 2 but at a much higher distance (about 12 units) than the other genotypes in the same group.
|
Estimates of cluster analysis under salinity conditions: Hierarchical cluster analysis based on the vegetative stage was presented in Fig. 8. Analyzing the dendrogram at distance (43), this study can observe two main distinct groups. The primary group comprises six genotypes with more substantial similarities: G46A, GAB-A, PR5, PR6, IR69625A/PR5 and IR69625A/PR6. Within the group, genotypes IR69625A/PR5 and IR69625A/PR6 show the closest relationship, joining at a low distance (4). Genotype PR6 joins this pair at a distance (11). While genotypes G46A and GAB-A show the closest relationship, joining at a low distance (5), genotype PR5 joins this pair at a distance (14). The secondary group encompasses the remaining 13 genotypes (Giza179, GZ9399, IR69625A/Giza179, IR69625A/GZ9399, G46A/GZ9399, GAB-A/GZ9399, GAB-A/PR6, G46A/Giza179, GAB-A/Giza179, GAB-A/PR5, G46A/PR6, G46A/PR5 and IR69625A). Analyzing the dendrogram, this second group can be divided into two distinct subgroups: Subgroup 1: This subgroup includes seven genotypes Giza179, GZ9399, IR69625A/Giza179, IR69625A/GZ9399, G46A/GZ9399, GAB-A/GZ9399 and GAB-A/PR6. Subgroup 2: This subgroup consists of six genotypes G46A/Giza179, GAB-A/Giza179, GAB-A/PR5, G46A/PR6, G46A/PR5 and IR69625A. Analyzing subgroup 1 divided into two sub-subgroups at a distance of about 15 to sub-subgroup 1, the group contains two genotypes, Giza179 and GZ9399. The sub-subgroup 2 contains five genotypes IR69625A/Giza179, IR69625A/GZ9399, G46A/GZ9399, GAB-A/GZ9399 and GAB-A/PR6. The sub-subgroup 1 showed that genotypes (G46A/GZ9399 and GAB-A/GZ9399) were the closest pair, joining at a distance of 6 and genotype GAB-A/PR6 joins the cluster at a distance of about eight units. In sub-subgroup 2, genotypes (IR69625A/Giza179 and IR69625A/GZ9399) were the closest pair, joining at a distance of 9. Analyzing subgroup 2 divided into two groups at a distance of about 22 to sub-subgroup 2 contains four genotypes G46A/Giza179, GAB-A/Giza179, GAB-A/PR5 and G46A/PR6. Sub-subgroup 2 showed that genotypes (GAB-A/Giza179 and GAB-A/PR5) were the closest pair, joining at a distance of 7 and genotype G46A/Giza179 joins the cluster at a distance of about nine units. Genotype IR69625A is somewhat more distinct within the group.
CONCLUSION
Finally based on this investigation the following genotypes could be used in the breeding program and expected improvement in such traits under normal and saline conditions, G46A, PR6, GZ9399, Giza179 and GAB-A as good donors. The G46A/PR6, G46A/PR5, GAB-A/ PR6 and IR69625A/Giza179 could be used as a good cross. The widespread exploitation of such promising hybrids will increase rice productivity under salt-affected soil conditions in the future.
SIGNIFICANCE STATEMENT
Salinity stress is the main grand challenge restricting rice production in Egypt, thereby investing the heterosis of hybrid rice to obtain high salt tolerance score and yield under such stress was amid. Using precise and correct statistical analysis showed high effectiveness in detecting some high salt tolerance hybrids, G46A/PR6, G46A/PR5, GAB-A/PR6 and IR69625A/Giza179 which was concomitant with high yield under salt stress too. The parental lines of tolerant hybrids could be recommended as donors of salt tolerance genes in rice breeding programs. Furthermore, the current statistical analysis could be recommended for use in the case of similar studies and conditions.
REFERENCES
- Omar, M.M., B.H.J. Massawe, M.J. Shitindi, O. Pedersen, J.L. Meliyo and K.G. Fue, 2024. Assessment of salt-affected soil in selected rice irrigation schemes in Tanzania: Understanding salt types for optimizing management approaches. Front. Soil Sci., 4.
- Negm, M.E., W. El-Kallawy and A.G. Hefeina, 2019. Comparative study on rice germination and seedling growth under salinity and drought stresses. Environ. Biodivers. Soil Secur., 3: 109-117.
- Zayed, B., S. Bassiouni, A. Okasha, M. Abdelhamed, S. Soltan and M. Negm, 2023. Path coefficient, eigenvalues, and genetic parameters in Egyptian rice (Oryza sativa L.) under aerobic conditions. SABRAO J. Breed. Genet., 55: 131-145.
- Ghosh, B., M.N. Ali and G. Saikat, 2016. Response of rice under salinity stress: A review update. J. Res. Rice, 4.
- Zayed, B.A., R.A. El-Namaky, S.E.M. Seedek and H.F. El-Mowafi, 2013. Exploration hybrid rice under saline soil conditions in Egypt. J. Plant Prod., 4: 1-13.
- Ramadan, E.A., M. Negm and M.A. Abdelrahman, 2020. Molecular analysis for salt tolerance QTLs emphasizing saltol QTL in some Egyptian and international rice genotypes (Oryza sativa L.). Egypt. Acad. J. Biol. Sci. C Physiol. Mol. Biol., 12: 57-69.
- Anis, G.B. and H.S. Gharib, 2016. Physical and physicochemical properties for selected hybrid rice combinations derived from three line system. J. Plant Prod., 7: 1155-1163.
- Wolf, B., 1982. A comprehensive system of leaf analyses and its use for diagnosing crop nutrient status. Commun. Soil Sci. Plant Anal., 13: 1035-1059.
- Bradford, G.R., H.D. Chapman, P.F. Pratt and A.P. Vanselow, 1961. Methods of Analysis for Soils, Plants, and Waters. University of California, Division of Agricultural Sciences, Oakland, California, Pages: 309.
- Flowers, T.J. and A.R. Yeo, 1986. Ion relations of plants under drought and salinity. Aust. J. Plant Physiol., 13: 75-91.
- Miller, P.A., J.C. Williams Jr., H.F. Robinson and R.E. Comstock, 1958. Estimates of genotypic and environmental variances and covariances in upland cotton and their implications in selection. Agron. J., 50: 126-131.
- Dewey, D.R. and K.H. Lu, 1959. A correlation and path-coefficient analysis of components of crested wheatgrass seed production. Agron. J., 51: 515-518.
- Jobson, J.D., 1992. Applied Multivariate Data Analysis: Volume II: Categorical and Multivariate Methods. 1st Edn., Springer, New York, ISBN: 978-1-4612-0921-8, Pages: 732.
- Negm, M.E., A.A. Hadifa, S.A. Soltan and S.M. Bassiouni, 2023. Genetic and molecular studies on some traits corresponding to salinity tolerance in rice. Menoufia J. Plant Prod., 8: 152-169.
- Zayed, B.A., I.S. El-Rafaee and S.E.M. Sedeek, 2010. Response of different rice varieties to phosphorous fertilizer under newly reclaimed saline soils. J. Plant Prod., 1: 1479-1493.
- Faiz, F.A., M. Sabar, T.H. Awan, M. Ijaz and Z. Manzoor, 2006. Heterosis and combining ability analysis in basmati rice hybrids. J. Anim. PIant Sci., 16: 56-59.
- El-Mowafi, H.F., M.D.F. AlKahtani, R.M. Abdallah, A.M. Reda and K.A. Attia et al., 2021. Combining ability and gene action for yield characteristics in novel aromatic cytoplasmic male sterile hybrid rice under water-stress conditions. Agriculture, 11.
- Sohrabi, M., M.Y. Rafii, M.M. Hanafi, A.S.N. Akmar and M.A. Latif, 2012. Genetic diversity of upland rice germplasm in Malaysia based on quantitative traits. Sci. World J., 2012.
- Osman, K.A., A.M. Mustafa, F. Ali, Z. Yonglain and Q. Fazhan, 2012. Genetic variability for yield and related attributes of upland rice genotypes in semi arid zone (Sudan). Afr. J. Agric. Res., 7: 4613-4619.
- Anis, G.B., A.I. Elsherif, T.M. Mazal, M.M. Osman and O.A. Elbadawy, 2024. Identification of new high yielding Indica-Japonica and effective restorer variety for different cytoplasmic male sterile (CMS) sources in rice. Menoufia J. Plant Prod., 9: 55-70.
- Akinwale, M.G., G. Gregorio, F. Nwilene, B.O. Akinyele, S.A. Ogunbayo and A.C. Odiyi, 2011. Heritability and correlation coefficient analysis for yield and its components in rice (Oryza sativa L.). Afr. J. Plant Sci., 5: 207-212.
- >Anis, G.B., 2019. Assessment of genetic variability and identification of fertility restoration genes Rf3, Rf4 of WA-CMS in RILs population of rice. J. Appl. Sci., 19: 199-209.
- Tripathi, N., O.P. Verma, P.K. Singh and P. Rajpoot, 2018. Studies on correlation and path coefficient analysis for yield and its components in rice (Oryza sativa L.) under salt affected soil. J. Pharmacogn. Phytochem., 7: 1626-1629.
- Abarshahr, M., B. Rabiei and H.S. Lahigi, 2011. Genetic variability, correlation and path analysis in rice under optimum and stress irrigation regimes. Notulae Sci. Biol., 3: 134-142.
- Kiani, S. and K. Agahi, 2016. Correlation and path coefficient analysis of morphological traits affecting grain shape in rice genotypes (Oryza sativa L.). Agric. For., 62: 227-237.
- Zarbafi, S.S., B. Rabiei, A.A. Ebadi and J.H. Ham, 2019. Statistical analysis of phenotypic traits of rice (Oryza sativa L.) related to grain yield under neck blast disease. J. Plant Dis. Prot., 126: 293-306.
- Zayed, B.A., W.H. El-Kellawy, A.M. Okasha and M.M. Abd El-Hamed, 2017. Improvement of salinity soil properties and rice productivity under different irrigation intervals and gypsum rates. J. Plant Prod., 8: 361-368.
How to Cite this paper?
APA-7 Style
Zayed,
B.A., Anis,
G.B., Abdel-Fattah,
A.G., Gharieb,
A.S., Elgamal,
A.A. (2025). Validation and Detection of Some New Rice Hybrids to Salt Stress Using Various Statistical Analysis Methods. Trends in Applied Sciences Research, 20(1), 1-13. https://doi.org/10.3923/tasr.2025.01.13
ACS Style
Zayed,
B.A.; Anis,
G.B.; Abdel-Fattah,
A.G.; Gharieb,
A.S.; Elgamal,
A.A. Validation and Detection of Some New Rice Hybrids to Salt Stress Using Various Statistical Analysis Methods. Trends Appl. Sci. Res 2025, 20, 1-13. https://doi.org/10.3923/tasr.2025.01.13
AMA Style
Zayed
BA, Anis
GB, Abdel-Fattah
AG, Gharieb
AS, Elgamal
AA. Validation and Detection of Some New Rice Hybrids to Salt Stress Using Various Statistical Analysis Methods. Trends in Applied Sciences Research. 2025; 20(1): 1-13. https://doi.org/10.3923/tasr.2025.01.13
Chicago/Turabian Style
Zayed, Bassiouni, A., Galal B. Anis, Abdel-Fattah G. Abdel-Fattah, Abdelfatah S. Gharieb, and Amgad A. Elgamal.
2025. "Validation and Detection of Some New Rice Hybrids to Salt Stress Using Various Statistical Analysis Methods" Trends in Applied Sciences Research 20, no. 1: 1-13. https://doi.org/10.3923/tasr.2025.01.13

This work is licensed under a Creative Commons Attribution 4.0 International License.


